PolyAffinity
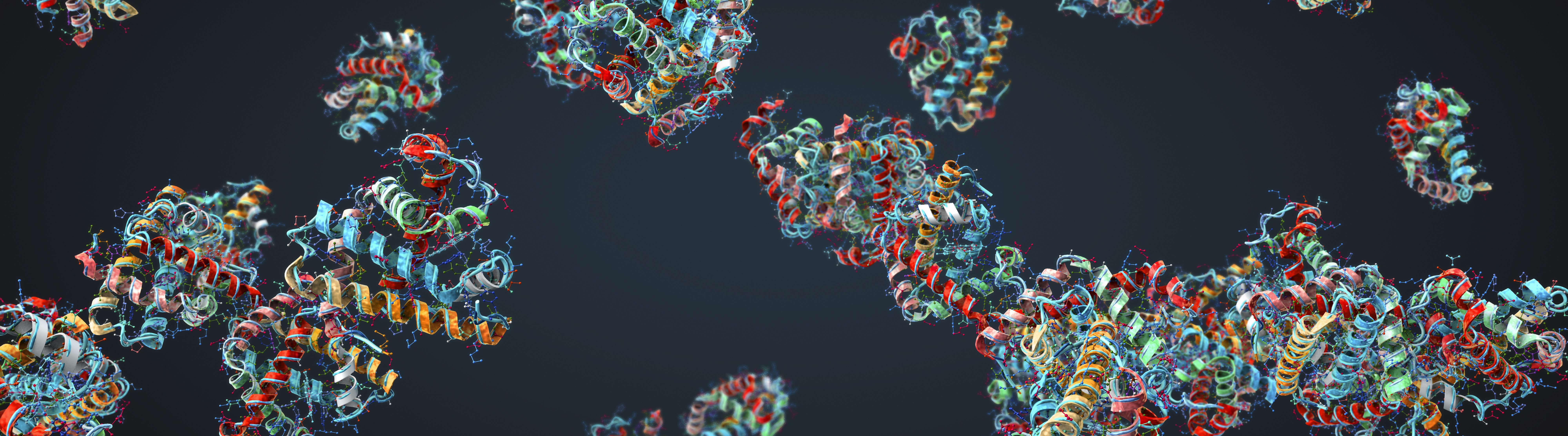

PolyAffinity aims to predict the binding affinity of protein-peptide complexes for traditionally hard-to-drug proteins, facilitating in-silco drug design where other tools fail. This is achieved using PolyChord's advanced-sampling technology to sample the energy landscape of the protein-peptide complex even in the presence of dynamic motion of the pair.
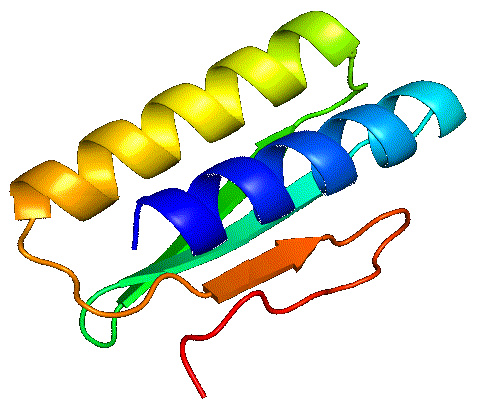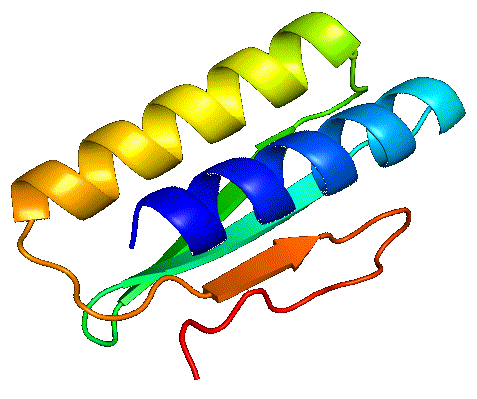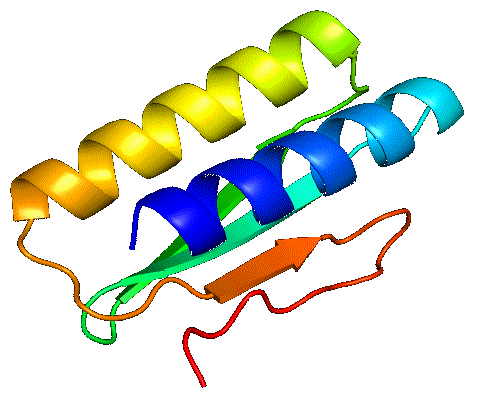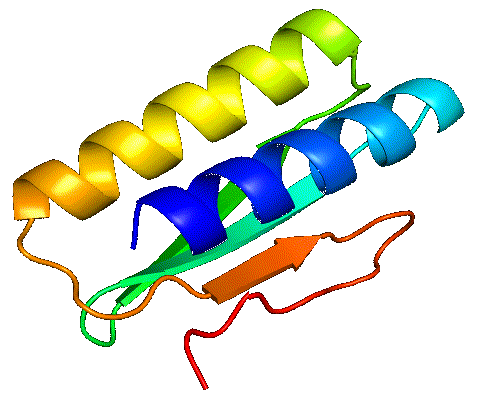
PolyAffinity's Protien Vibration Visualisation
Traditional machine-learning-based methods have been shown to be very effective for predicting protein structure, but they essentially provide a snapshot “photograph” of the protein. In reality, a protein is dynamically moving, or vibrating, and in many cases (for example, with so-called disordered proteins) it is important to account for this dynamic motion in making predictions. By fully sampling the energy surface PolyAffinity can account for this.
By fully sampling the energy landscape of the complex, PolyAffinity is also able to identify alternative states to the native one, and account for them in its prediction of binding affinity. Furthermore, the prediction's temperature can be adjusted post-analysis to examine how the protein-peptide complex behaves in different environmental conditions.
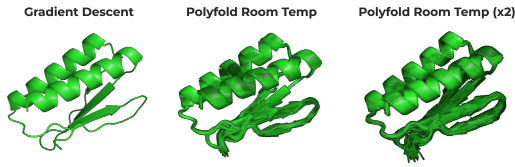
PolyFold Capturing Vibrational Motion at Any Temperature

PolyFold's Navigation of Full-Atom Energy Surface
This exact same method can also be used in refining the structure of individual proteins in a way that accounts for dynamic motion, misfolds and alternative states. This is just a taste of what PolyAfinity can achieve. To learn more, head to our contact us page and shoot us a message for more details.